Data Schemas
Data schemas play a crucial role in organizing information effectively by offering precise definitions of entities and their relationships. These structured forms not only facilitate data indexing but also enable seamless metadata export. As valuable assets, these schemas significantly contribute to enhancing data findability on the web. Moreover, they play a pivotal role in improving machine-readability and facilitating data analysis. A noteworthy aspect is that many of these schemas support the utilization of ontologies. They achieve this by either accepting or requesting values to be provided as ontology terms and by providing links and identifiers to these terms, further enhancing the depth and richness of the structured data.
nmrXiv Data Model
nmrXiv initially adopted the ISA (Investigation, Study, Assay) approach for structuring experimental metadata, as outlined by the ISA specifications. However, nmrXiv has chosen a distinct terminology to align with its specific requirements:
- ISA Investigation ⇛ nmrXiv Project
- ISA Study ⇛ nmrXiv Study
- ISA Assay ⇛ nmrXiv Dataset
For a comprehensive understanding of the objects and their relationships, you can refer to the Data Model folder.
As each study corresponds to one sample and each dataset corresponds to one spectrum, nmrXiv frontend tends to display these later, more chemistry-friendly terms to enhance user experience.
In a recent development, nmrXiv has extended its data model to accommodate the submission of individual samples independently, eliminating the requirement to include them in projects. This enhancement aims to streamline the submission process, reducing complexity and offering users greater flexibility when submitting single samples to the platform.
Data Schemas in nmrXiv
nmrXiv's emphasis on obtaining Digital Object Identifiers (DOIs) for various entities, coupled with a commitment to describe the data using ontology terms, has prompted the prioritization of a list of schemas. While some schemas have already been implemented, others are in the pipeline and will be introduced soon. Below is a list of these schemas:
Bioschemas (Repository, Project, Studay, Dataset, Sample, Molecule)
Bioschemas aims to improve the Findability of life sciences resources (such as datasets) on the Web by using Schema.org markup in the websites, so that they are indexable by search engines and other services. Bioschemas is making two main contributions:
- Proposing new types and properties to Schema.org to allow for the description of life science resources.
- Defining usage profiles over the Schema.org types that identify the essential properties to use in describing a resource.[1]
nmrXiv uses Bioschemas types to describe both data structuring components (such as datasets) and experiment-related components (such as samples and molecules). Here is a list of the used types:
DataCatalog
The DataCatalog type can be used to describe repositories, and thus, it is used to describe nmrXiv repository.DataCatalog API endpoint link
Study
While the Schema.org already encompasses a Project type, it primarily pertains to enterprises rather than scientific investigations. Given the similarity in metadata structure between project and study in nmrXiv (e.g., id, name, URL), nmrXiv leverages the study draft offered by Bioschemas to represent projects. This enables seamless linking to included studies and their respective datasets. Project API endpoint link.
Moreover, nmrXiv adopts the study draft to describe the study. The properties of this draft can establish connections between the study, its parent project, its associated sample, and the contained datasets. Importantly, this study entity is an integral part of a broader project. Study API endpoint link.
ChemicalSubstance
The ChemicalSubstance type is used in nmrXiv to characterize NMR samples. It is associated with the study through the "about" property, indicating that the study pertains to this specific sample. Additionally, the molecules present in the sample are incorporated into it as values of the "hasBioChemEntityPart" property. While there is no dedicated API endpoint for the Sample, it can be accessed within the Study.
MolecularEntity
The MolecularEntity erves to characterize any constitutionally or isotopically distinct atom, molecule, ion, etc., identifiable as a separately distinguishable entity. Currently, its application in nmrXiv is focused on describing the molecules present in a sample. However, it holds potential for broader applications, such as detailing the nucleus or the solvent. While there is no dedicated API endpoint for the MolecularEntity, it is accessible within the Study ⇛ Sample structure.
Dataset
The Dataset schema is used in nmrXiv to describe the datasets there. Each dataset is linked to its parent project and study through the "isPartOf" property. Dataset API endpoint link.
ISA (Sample, Assay, Ontology)
ISA is a metadata framework to manage a diverse set of life science, environmental, and biomedical experiments that employ one or a combination of technologies, built around the Investigation (the project context), Study (a unit of research), and Assay (analytical measurement) data model and serializations (tabular, JSON and RDF).[2]
nmrXiv will comply with ISA Schema specifications to capture NMR metadata (Assay, Sample, and Ontology) to provide a detailed description of the experimental metadata for both synthetic and biological experiments (i.e., sample characteristics, technology and measurement types, sample-to-data relationships). ISA enables interoperability with other platforms and services while keeping the information in a simplistic and open-text format that is FAIR-compliant. nmrXiv back-end will be designed to support ISA for newly entered data. However, ISA still comes with few limitations from the data management (repository) perspective, configurable templates on the go and redundant data being some of them. nmrXiv will contribute to extending the existing ISA models with NMR domain-specific requirements to ensure total flexibility for the end user to define their own templates while still being complaint with ISA Specifications and the minimum information standards.
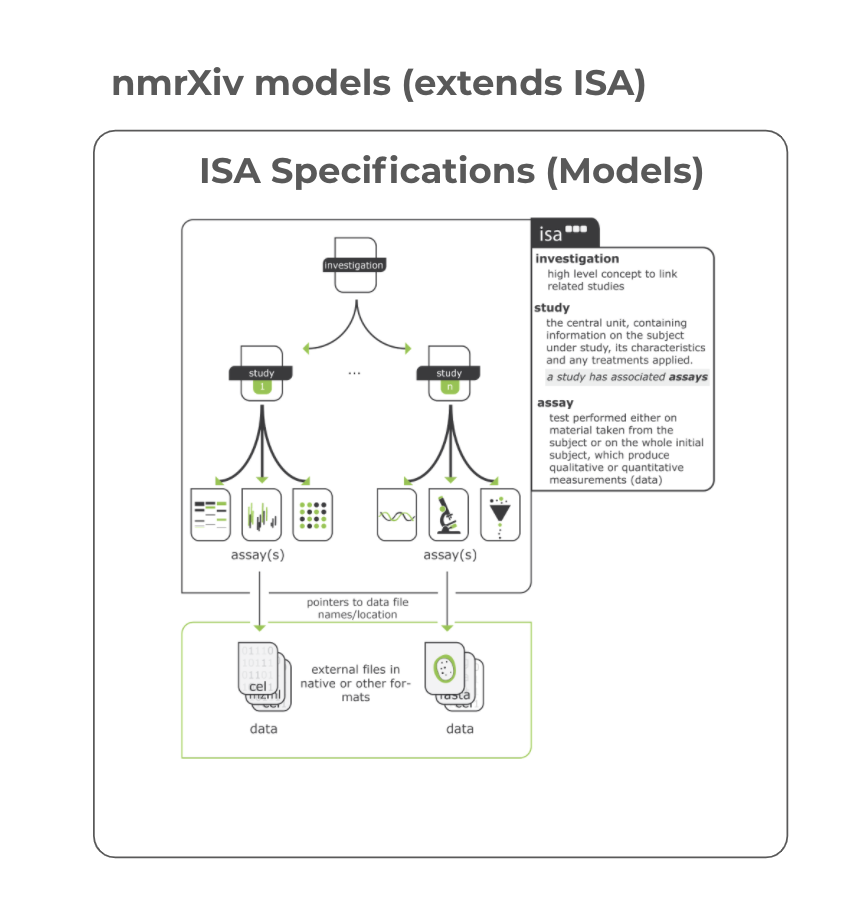
ISA Sample Schema
In a Study object, ISA records the provenance of biological samples from source material through a collection process to sample material represented with directed acyclic graphs (direct graphs with no loops/cycles). The pattern of nodes is usually formed of a source material node, followed by a sample collection process node, followed by a sample material node.
(source material)->(sample collection)->(sample material)
These study graphs MAY split and pool depending on how the samples are collected.
In a splitting example, multiple samples might be derived from the same source:
(source material 1)->(sample collection)->(sample material 1)
(source material 1)->(sample collection)->(sample material 2)
In a pooling example, multiple sources may be used to create a single sample:
(source material 1)->(sample collection)->(sample material 1)
(source material 2)->(sample collection)->(sample material 1)
However, sample collection applies only to biological samples where the source comes from a certain organism, while with synthetic samples we have a certain compound that gets dissolved in a solvent to create the sample. We are trying to capture this concept in ISA adopting mock-ups derived from actual samples/molecules found in non-biological repositories such as Chemotion and NMRShiftDB, and compare them with a biological sample coming from the biological repository MetaboLights. Please find the mentioned mock-ups here. Those mock-ups are still under development, especially due to the absence of the relevant ISA configurations in addition to the continuous update upon discussion.
nmrXiv approach to represent a synthetic sample is to take the compound found in Chemotion or NMRShiftDB as a source, and apply an NMR sample protocol where a solvent gets added to generate the sample, and where the solvent becomes one of the sample characteristics. This NMR sample undergoes an NMR spectroscopy protocol with parameters such as the instrument, magnetic field strength, pulse sequence name, temperature and others. This protocol will result in a data file, which, in return, undergoes an NMR assay protocol with some NMR processing software. This approach is still under discussion and not confirmed.
However, we are still facing issues regarding:
- Missing ontology terms to represent the characteristics and factors categories.
- The limited use of ontologies in both Chemotion and NMRShiftDB, so we picked terms representing the fields and values in the mock-ups despite the frequent use of free text in the actual models.
- Representing characteristics and factors categories when the category is a combination of two or more ontology terms. e.g., "number of rings", "duration of incubation".
ISA Ontology Schema
ISA ontology reference schema can capture ontology terms where those "annotation_values" are linked to their "term_source" which is the ontology the term comes from, and the "term_accession", which is the link to that term.
ISA Converters
nmrXiv will contribute to developing data converters as independent microservices (python-based applications) to convert data from and to ISA, which enables nmrXiv and other repositories to utilize these converters as plugins to capture and export data. nmrXiv will also leverage the existing tool set in the domain, such as nmrml2isa converter, to capture rich metadata from the open nmrML files. nmrml2isa is a Python3 program that can be used to generate an ISA-Tab structured investigation out of nmrML files, providing the backbone of a study that can then be edited further to provide additional metadata that cannot be automatically extracted.
IUPAC - FAIRSpec (Spectra)
nmrXiv will contribute to and adopt the definition of standardized metadata being developed within IUPAC Project "Development of a Standard for FAIR Data Management for Spectroscopic Data", which covers spectroscopic data including NMR spectroscopy. But this project is still preliminary and under development.
DataCite (Data Citation and Discovery)
The DataCite Metadata Schema is a list of core metadata properties chosen for accurate and consistent identification of a resource for citation and retrieval purposes, along with recommended use instructions. It primarily supports citation of data rather than discipline-specific metadata.
Complying with the DataCite schema enables the creation of new Digital Object Identifiers (DOIs) and their assignment to content on nmrXiv, which is essential for identifiers' persistence. Only mandatory property values are needed to obtain the DOI, but still, there is the ability of rich citing-metadata description with recommended and optional properties. nmrXiv will mostly treat some recommended fields as mandatory since they are expected to become mandatory in DataCite in the near future and to make sure all essential metadata is captured. Mandatory fields include the DOI, Creator, Title, Publisher, PublicationYear, and ResourceType. Additionally, using DataCite schema will enable easy export of the metadata in XML format.
OpenAIRE Guidelines (Data Citation and Discovery)
Complying with OpenAIRE Guidelines for Data Archive Managers ensures compatibility with the OpenAIRE infrastructure, which facilitates interoperability with other repositories already adhering to those guidelines, enhancing data exposure and visibility. OpenAIRE has already adopted the DataCite Metadata Schema, yet with some minor adjustments, such as accepting other persistent identifier schemes rather than the DOI and some changes in the obligations of properties.

